|
Chapter F – Ms/ms search of two proteins.
1 – Preparations.
The background for the analysis is and analysis of the interaction of alpha-1-antitrypsin and proteinase 3. A gel-band is observed at the position of PR3, but it is suspected that it may be a-1-a. Instead of just analysing by MALDI, we also want to have as good a sequence coverage as possible, so we analyse it by ms/ms using an Orbitrap XL after tryptic digestion.
We want to use the X!Tandem search engine of GPMAW, and as there are only two proteins involved, there is no need to search the large database.
 Search data: The data are collected by a short 30 min separation using nanoLC, and the data are converted into a peak list. An mgf file format is chosen as this contains a bit more information than dta or pkl. The data are saved in a separate directory. Search data: The data are collected by a short 30 min separation using nanoLC, and the data are converted into a peak list. An mgf file format is chosen as this contains a bit more information than dta or pkl. The data are saved in a separate directory.
Getting the proteins: The accession number of each of the two proteins (AAH96186 and P01009) are entered in main toolbar retrieval box and the sequences retrieved from the web.
Saving the proteins: As X!Tandem reads FastA formatted files, the two sequences are saved to file by selecting File | Export sequence | all sequences as FastA file. The file is saved with the name PR3A1A.TXT in the same directory as the ms/ms data to keep the project organized.
Setting up the search: The ms/ms search dialog is called through Search | MS/MS Search (an alternative would be pressing the F5 key or the shortcut in the toolbar).
The files created are now selected either through the ‘Open’ buttons, or simply by drag-and-drop from File Explorer.
The enzyme chosen is trypsin, the output file name is left at default, the ‘Search crap list’ (contaminations) is checked.
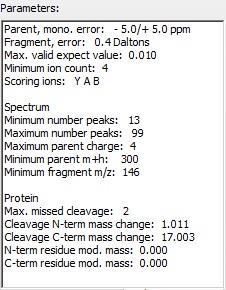 The parameters are chosen to suit the Orbitrap XL, in particular precision is set at 5 ppm (an initial search of a previous file had shown that the instrument was properly calibrated), fragment error at 0.4 Da, e-value at 0.01, maximum charge at 4 and missed cleavage at 2. The parameters are chosen to suit the Orbitrap XL, in particular precision is set at 5 ppm (an initial search of a previous file had shown that the instrument was properly calibrated), fragment error at 0.4 Da, e-value at 0.01, maximum charge at 4 and missed cleavage at 2.
As the search is quite fast (on my machine 3 sec) you can play around with the parameters to get the best results. If you do not change the ‘output file name’ the result files will overwrite each other and you will not clutter up your disk with temporary data. If you want to save your results, change the output file name when changing parameters. When you load a new search file, the name will be changed automatically.
Oxidation of Met and Carbamidomethyl modification of Cys are chosen as variable and fixed modifications.
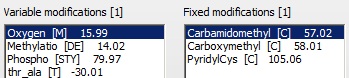
‘Enable refinement’ is chosen, with ‘Semi cleavage’ checked, as we may find unusual cleavages (i.e. the gel band is at a lower Mr than expected for a-1-a).
2 – Results.
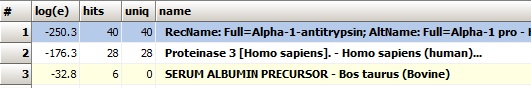
 The results of the search show that alpha-1-antitrypsin is likely to be the major component in the gel band analyzed. The results of the search show that alpha-1-antitrypsin is likely to be the major component in the gel band analyzed.
The precision of the parent ions are quite good, although the calibration is not perfect (between -4 and 0 ppm for Alpha-1-antitrypsin, see right).
Alpha-1-antitrypsin is the first hit with the major score, so it is clear that the gel-band contains a major proportion of this protein. A little surprisingly bovine serum albumin is also on the hit list, so now the search goes in for finding the contamination.
Double clicking on the first line with alpha-1-a, fills out the result page and displays the sequence with identified peptides shown in red.
 From the annotation page of the sequence the reported signal sequence is 1-24, which fits perfectly with the first peptide starting at residue 25. However, comparing with runs of tryptic digests of the intact protein, a large number of peptides are missing up to residue 150. If the protein started at residue 150 or slightly earlier, the resulting mass of the protein would be just over 30 kDa, which would fit with the relative molecular weight observed in the gel. The reason for the first peptide to be observed may be caused by the presence of three histidines, giving the peptide high proton affinity and thus making it easy to observe. From the annotation page of the sequence the reported signal sequence is 1-24, which fits perfectly with the first peptide starting at residue 25. However, comparing with runs of tryptic digests of the intact protein, a large number of peptides are missing up to residue 150. If the protein started at residue 150 or slightly earlier, the resulting mass of the protein would be just over 30 kDa, which would fit with the relative molecular weight observed in the gel. The reason for the first peptide to be observed may be caused by the presence of three histidines, giving the peptide high proton affinity and thus making it easy to observe.
The ms/ms spectrum of the 25-49 peptide is not convincing, having low intensity and the major peaks not identified (below right),
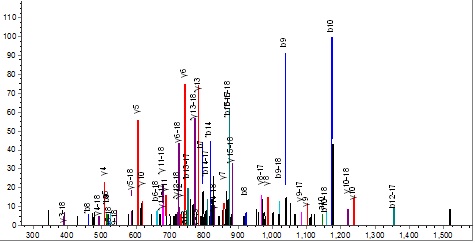
however, the 35-49 peptide is difficult to refuse.
Clicking on the ‘Sequence’ button  opens the coverage window, which shows a detailed picture of the peptides identified. opens the coverage window, which shows a detailed picture of the peptides identified.
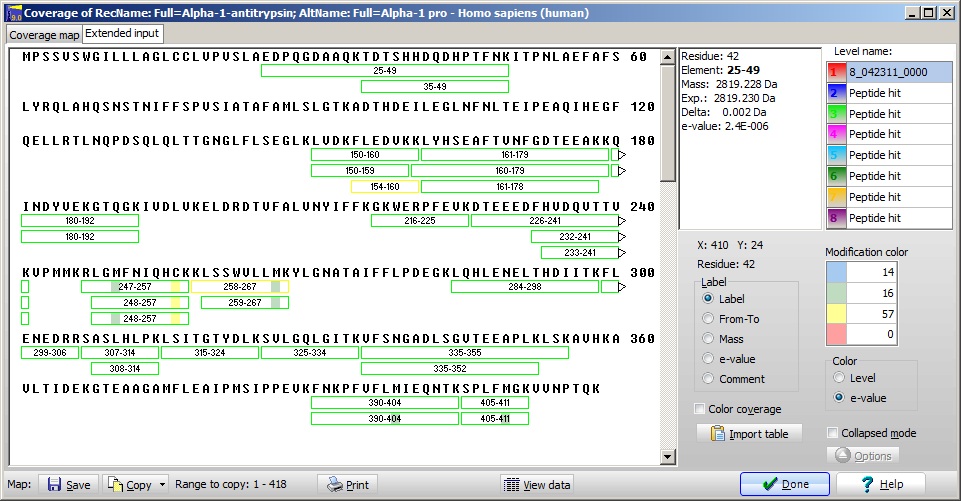
The conclusion must be that we most likely have a truncated form of alpha-1-antitrypsin cleaved at residue 150 or slightly before, but it is ‘contaminated’ by a small amount of full-length protein which demands further analysis.
Note: When you perform a number of ms/ms searches, the results are saved in xml files in the search directory. These can be re-opened through the ‘Open result file’ button, or more easily by drag-and-drop from the File Explorer. If you want to compare with another search directly, you can open a second search window and load the result file into this.
|