|
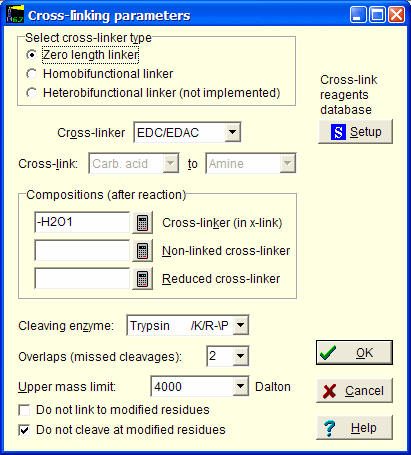
In this dialog you first select cross-linker type.
Depending on the selection, the cross-linker drop-down box will fill with the appropriate linkers available in the local database.
If you cannot find your cross-linker here, click on the Setup button to edit the database.
The cross-link from to is set by the database.
Compositions are set by the database, but can be edited.
Finally you select the enzyme selected for cleavage of the protein(s), number of missed cleavage and the upper mass limit you want to analyze.
The enzyme you can select are from the standard list available in “Automatic digest”
|